|
The epitranscriptome in infectious disease and cancer
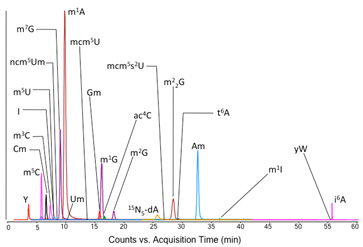
 A major new line of research addresses the role of RNA secondary modifications in the cellular responses to toxic exposures. Over 100 different ribonucleoside structures have been identified in both prokaryotes and eukaryotes, most prominently in tRNA and rRNA, with ~20-30 types of modifications present in any one organism. Recent studies suggest an important role for the various nucleoside structures in controlling translation in response to cell stimuli. We have developed a sensitive LC-MS/MS approach to identifying all of the RNA modifications in an organism and quantifying changes in the spectrum of modifications following cellular exposures. A typical composite chromatogram is shown in the adjacent figure, with an example of a PCA analysis of changes in the modification A major new line of research addresses the role of RNA secondary modifications in the cellular responses to toxic exposures. Over 100 different ribonucleoside structures have been identified in both prokaryotes and eukaryotes, most prominently in tRNA and rRNA, with ~20-30 types of modifications present in any one organism. Recent studies suggest an important role for the various nucleoside structures in controlling translation in response to cell stimuli. We have developed a sensitive LC-MS/MS approach to identifying all of the RNA modifications in an organism and quantifying changes in the spectrum of modifications following cellular exposures. A typical composite chromatogram is shown in the adjacent figure, with an example of a PCA analysis of changes in the modification  spectrum following exposure to hydrogen peroxide and other toxicants. This “top down” approach has led to the identification of RNA modifications and their biosynthetic pathways that are critical to the cell survival response. spectrum following exposure to hydrogen peroxide and other toxicants. This “top down” approach has led to the identification of RNA modifications and their biosynthetic pathways that are critical to the cell survival response.
As part of the Infectious Disease group of the Singapore-MIT Alliance for Research and Technology, we have embarked on a study of RNA modifications in tuberculosis and malaria. The goal is to characterize the role of RNA modifications, starting with tRNA, in the response of microbial pathogens to reactive oxygen and nitrogen species generated by phagocytes in the human immune response.
|
