|
The epigenetics of the human gut microbiome and microbial pathogens
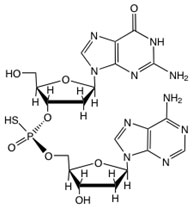 Phosphorothioate (PT) modification of the DNA backbone involves replacement of a non-bridging phosphate oxygen with a sulfur atom. Following the pioneering work of Eckstein and coworkers, PT modification of DNA has long been known as a synthetic modification that stabilizes oligodeoxynucleotides against nuclease degradation. However, we recently worked with the Deng group at Shanghai Jiao Tong University in our discovery of PT modifications as natural products in DNA from bacteria harboring the five-gene dnd cluster, with the PT modification located in G-G and G-A sequence contexts in Streptomyces lividans and E. coli B7A, respectively. The discovery of sequence-selective PT modification of DNA raises many questions about the function of this enzymatically-mediated, post-synthetic modification of DNA in cell physiology. Is it part of a restriction-modification system? What are the broader sequence and genome contexts for PT? How widespread are the dnd genes responsible for the modification?To address these issues, we have undertaken a series of studies to define the quantity, location and function of PT modifications of DNA in bacterial genomes. Phosphorothioate (PT) modification of the DNA backbone involves replacement of a non-bridging phosphate oxygen with a sulfur atom. Following the pioneering work of Eckstein and coworkers, PT modification of DNA has long been known as a synthetic modification that stabilizes oligodeoxynucleotides against nuclease degradation. However, we recently worked with the Deng group at Shanghai Jiao Tong University in our discovery of PT modifications as natural products in DNA from bacteria harboring the five-gene dnd cluster, with the PT modification located in G-G and G-A sequence contexts in Streptomyces lividans and E. coli B7A, respectively. The discovery of sequence-selective PT modification of DNA raises many questions about the function of this enzymatically-mediated, post-synthetic modification of DNA in cell physiology. Is it part of a restriction-modification system? What are the broader sequence and genome contexts for PT? How widespread are the dnd genes responsible for the modification?To address these issues, we have undertaken a series of studies to define the quantity, location and function of PT modifications of DNA in bacterial genomes. 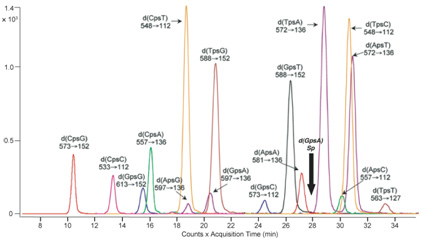 Our initial efforts led to the development of an LC-MS/MS method that screen for the presence of PT in genomic DNA, to identify the two-nucleotide sequence context (of 16 possible) and to quantify PT. A representative chromatogram is shown in the adjacent figure. The application of this method revealed that sequence-specific PT modifications are widespread in bacterial communities and are quantized at three distinct levels that are consistent with the DNA sequence recognition elements of other DNA modification systems. Our initial efforts led to the development of an LC-MS/MS method that screen for the presence of PT in genomic DNA, to identify the two-nucleotide sequence context (of 16 possible) and to quantify PT. A representative chromatogram is shown in the adjacent figure. The application of this method revealed that sequence-specific PT modifications are widespread in bacterial communities and are quantized at three distinct levels that are consistent with the DNA sequence recognition elements of other DNA modification systems.
|
